Usage of PrDOS
Prediction Submission
Input protein amino acid sequences in plain text or FASTA format into the form.
Multiple FASTA formatted inputs are acceptable.
The number of sequences in the multiple FASTA formatted input is limited to 50,
due to the limitation of the computational resources.
The server accepts the 20 single letter codes for standard amino acids and
the code 'X' generally used for non-standard amino acids.
The server automatically replaces other codes for ambiguous amino acids
and paticular non-standard amino acids by 'X',
and removes whitespaces in the query sequence.
Too long sequences with lengths greater than 2000 residues are not accepted
because of calculation costs.
| Amino acid | Single letter code |
|---|
| Alanine | A |
| Cysteine | C |
| Aspartic Acid | D |
| Glutamic Acid | E |
| Phenylalanine | F |
| Glycine | G |
| Histidine | H |
| Isoleucine | I |
| Lysine | K |
| Leucine | L |
| Methionine | M |
| Asparagine | N |
| Proline | P |
| Glutamine | Q |
| Arginine | R |
| Serine | S |
| Threonine | T |
| Valine | V |
| Tryptophan | W |
| Tyrosine | Y |
| Amino acid | Single letter code |
|---|
| Asparagine or aspartic acid | B |
| Glutamine or glutamic acid | Z |
| Leucine or Isoleucine | J |
| Amino acid | Single letter code |
|---|
| Selenocystein | U |
| Pyrrolysine | O |
There is trade-off between prediction sensitivity (true positive rate) and false positive rate.
If you permit higher false positive rate, you can obtain more sensitive prediction.
The acceptable false positive rate or expected sensitivity depends on the aim
of predictions.
The receiver-operating characteristic (ROC) curve shows
the sensitivity (true positive rate) for a particular false positive rate.
If the user want to recover at least 60% of disordered regions,
the user should set false positive threshold at 4%.
The default false positive rate is set at 5%.
ROC curves of PrDOS
The prediction system is composed of two predictors
(
see the detail).
The template prediction is useless when the user wants to know just
the disorder tendency of local amino acid composition.
The user can predict disordered regions without template prediction by this option.
If "Recieve prediction results by e-mail" checkbox is not checked,
this page will show a prediction progress report until all prediction processes are finished,
and finally return HTML formatted prediction results.
Although it depends on the length of query protein and server conditions,
PrDOS usually takes from 5 to 10 minutes to predict a protein sequence.
Thus, using "Recieve prediction results by e-mail" option is strongly recommended.
E-mail results include a link to the same HTML formatted prediction results.
Output format of prediction results
E-mail outputs
Prediction results returned by e-mail include a link to HTML formatted prediction results
and predicted disorder probability of each residue in plain text format.
The prediction results are composed of 4 columns.
The first column is a residue number, and the second is an amino acid type of the residue.
If '*' in the third column, the residue is predicted disordered.
Final column shows disorder probability of the residue.
Example of e-mail outputs
No AA Pred Probability
--------------------------
1 A * 0.85
2 W * 0.76
3 L * 0.82
4 E * 0.84
5 A * 0.82
6 Q * 0.79
7 E * 0.80
8 E * 0.79
9 E * 0.75
10 E 0.68
11 V 0.61
12 G 0.53
HTML formatted outputs
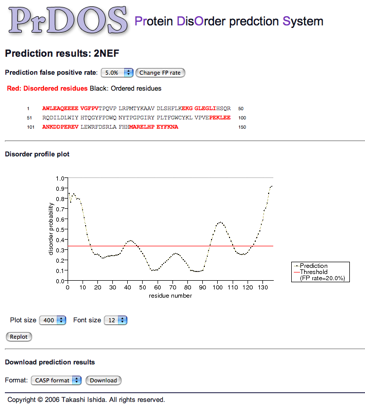
The prediction result page is consisted of three parts. The top part shows the prediction result of the two-state prediction (disorder/order). The red residues are predicted to be disordered at the given prediction false positive rate. The middle part shows the plot of disorder probability of each residue along the sequences. Residues beyond the red threshold line in this plot are predicted to be disordered. The user can change the size of the plot through the web-interface. At the bottom part, the user can download the raw prediction results in CSV format or CASP format.
Example of HTML formatted outputs